OvalY - Codon to Structure Investigation
Introduction
The Alphafold website provides protein bonding predictions for many proteins. Each has amino acid locations and confidence levels. There are also structural annotations.
I found the OvalY protein by looking for high-confidence estimates to see how the shape corresponds to the codon patterns used to code each aminio acid.
Here’s an overview of how the major features of OvalY compare with their codon patterns.
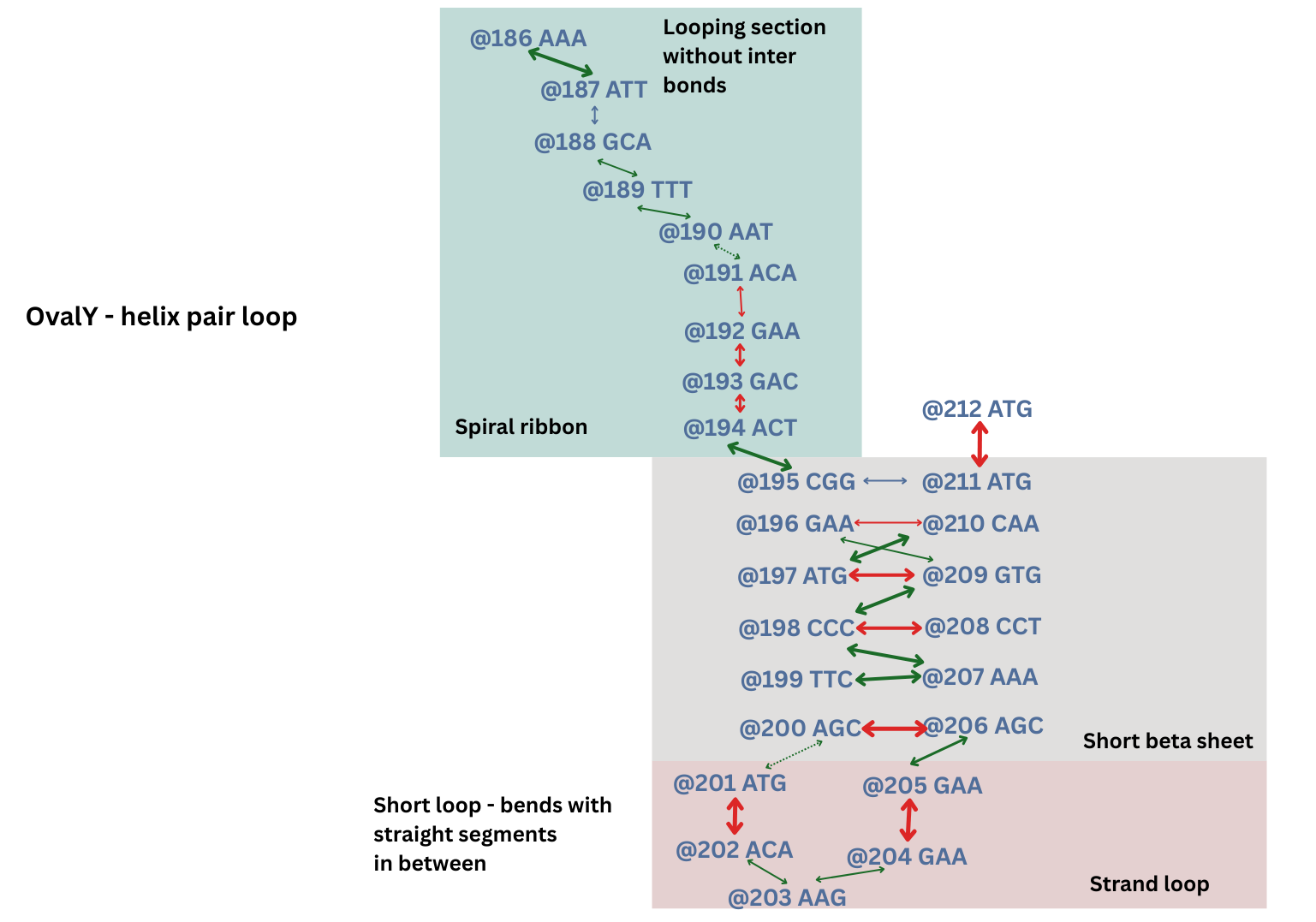
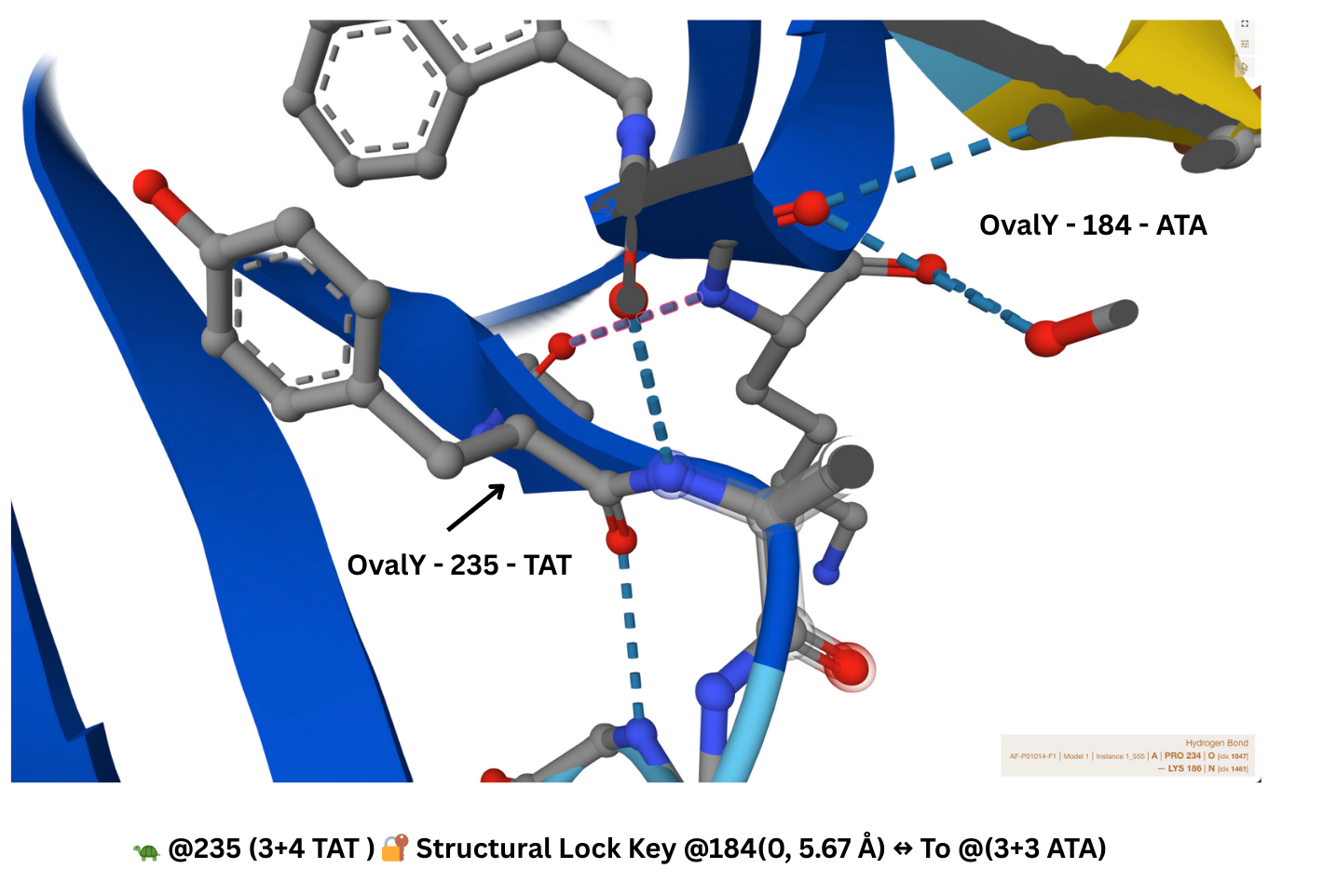
Lock and Key Site
One of the major junctions is a bonding site where Alpha Fold shows bonds from 184-236 and 186-236. Looking at the image though, the closest two residues are 184 and 235 where we have TAT and ATA - perfect opposite signatures. Also, there are compatible amino acid shapes. TAT’s has Tyrosine - a phenol ring and ATA has Isoleucine - a long branch that bonds with the ring. The genetic analyzer shows the distance between the two as 5.67 Å.
Flow Bonding Site on Small Beta Sheet
A protein beta sheet are opposite strands that link up:
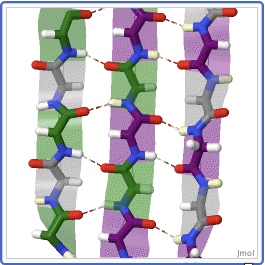
“Proteopedia, Life in 3D”. proteopedia.org.
Here’s a small beta sheet region from Alphafold showing a bond region where ATGs at 212 and 214 bond with 282:
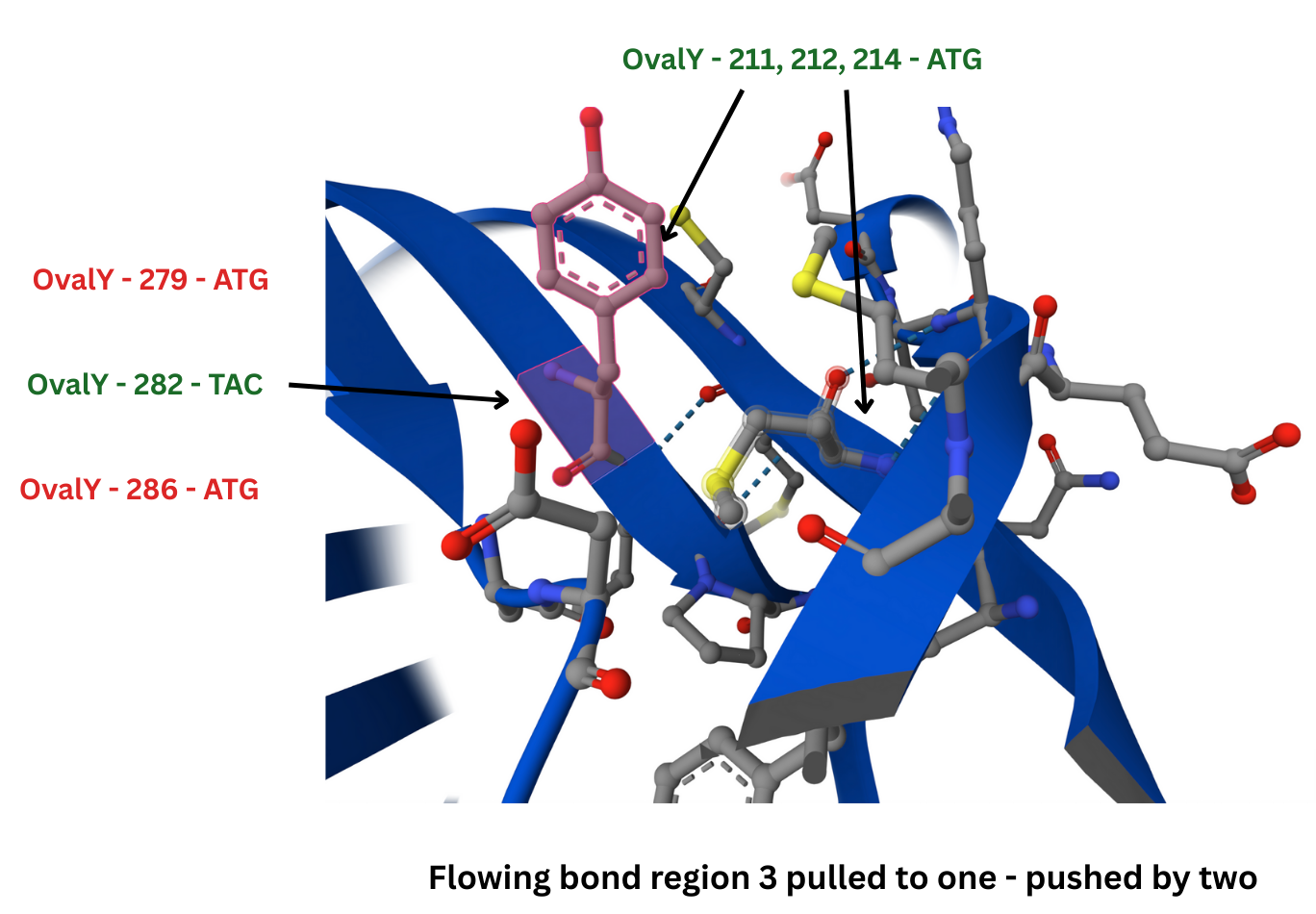
and here is that small sheet showing the bond strengths:
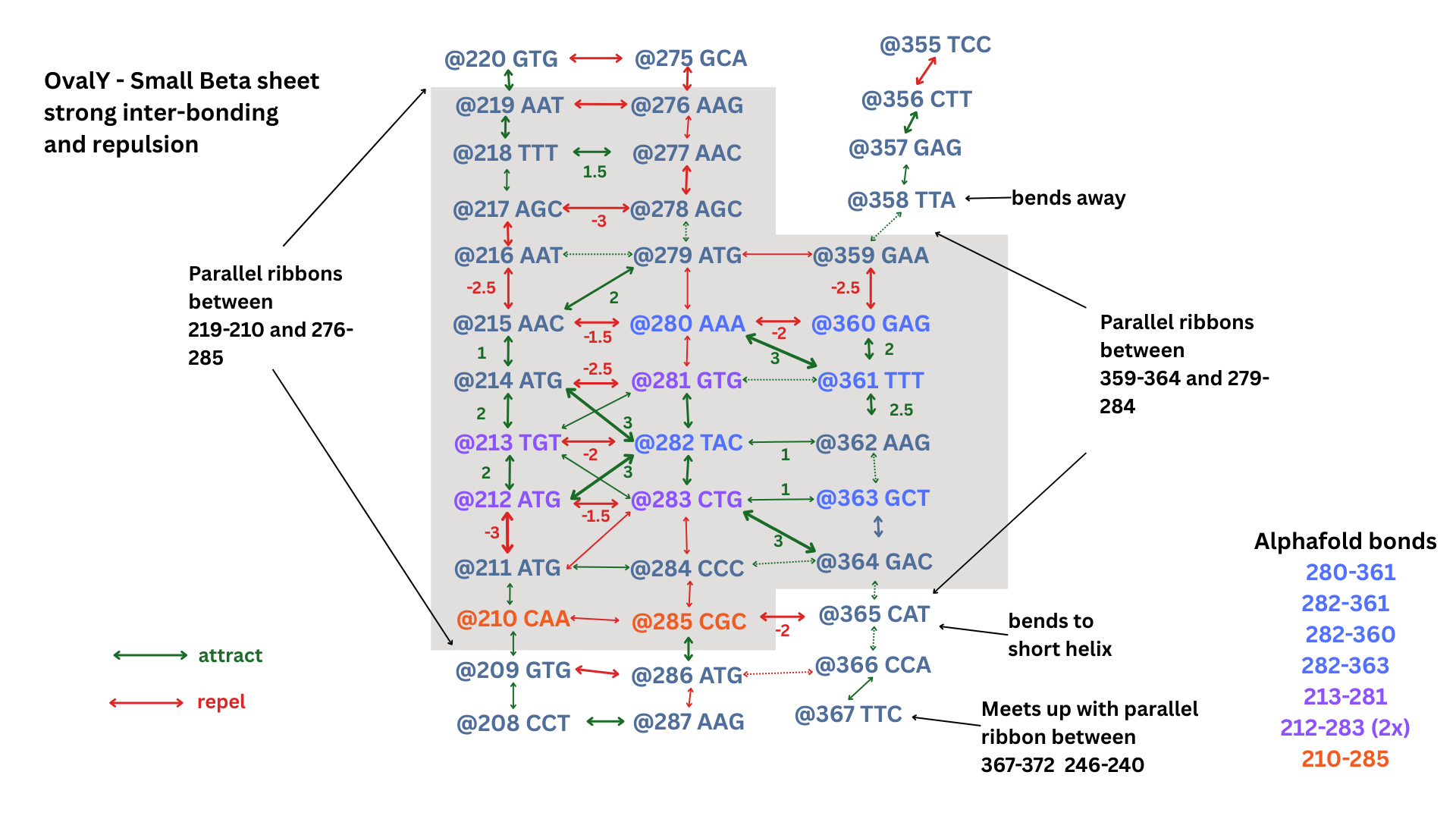
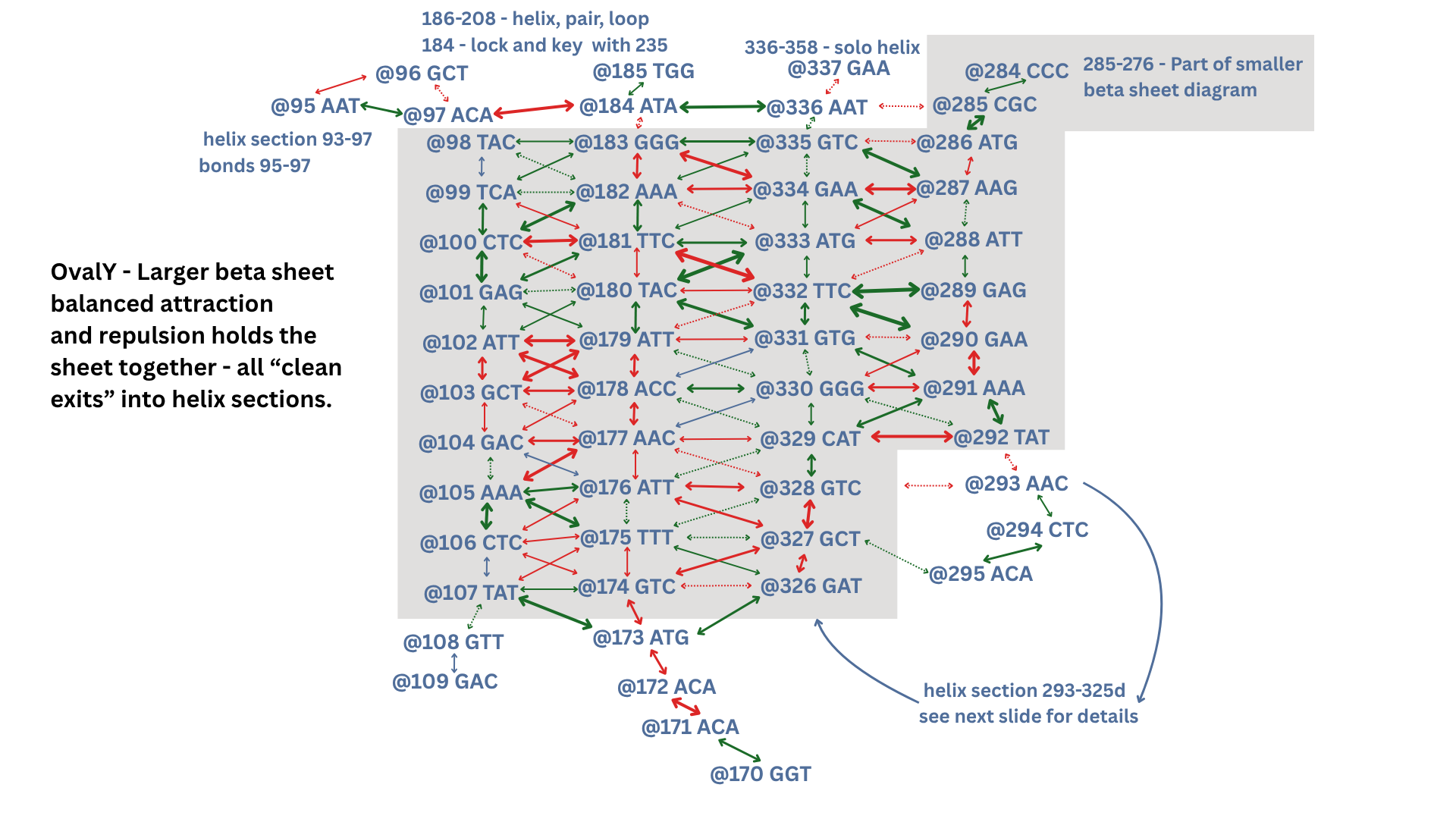
Larger Beta Sheet
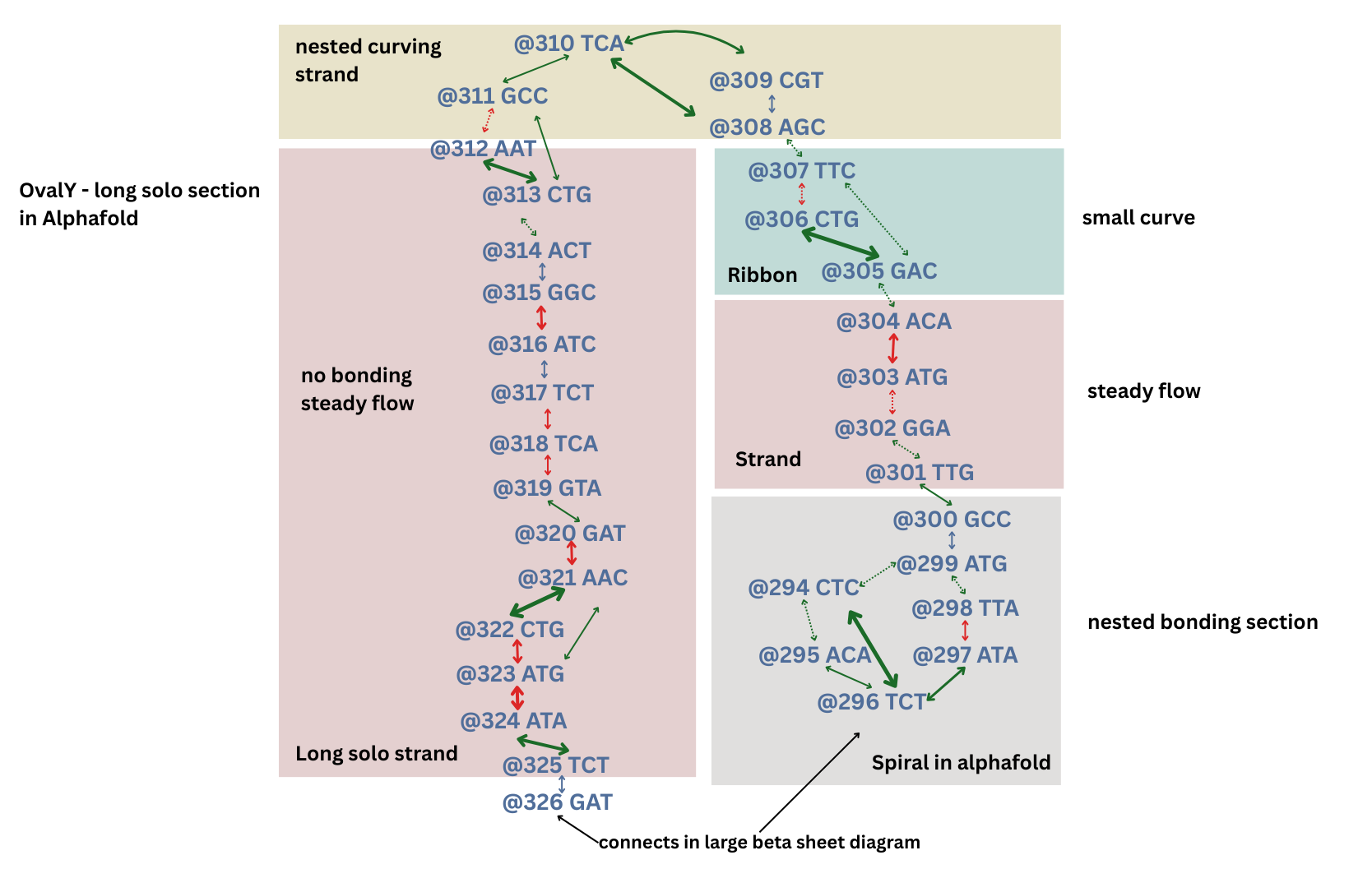
Long solo section
Helix pair loop